Cytochrome oxidaseComplex IV |
Cytochrome oxidase is one of a superfamily of proteins which act as the terminal enzymes of respiratory chains. The two main classes are cytochrome c oxidases, and quinol oxidases. The common features are:
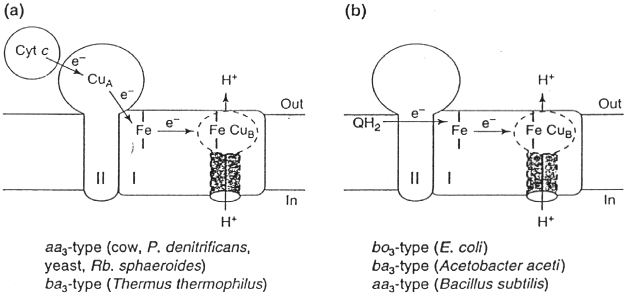
Some of the variations among the heme-Cu oxidase superfamily are shown in this scheme. Cytochrome c oxidases (left) of mitochondria, of respiratory bacteria from the purple bacterial branch of the Eubacteria, and one of the cytochrome c oxidases of Rhodobacter sphaeroides, are of the aa3-type. Others (ba3-type) have a heme b in place of heme a. Not shown are cytochrome c oxidases which have cytochrome b attached to subunit II, and cytochrome oxidases (cbb3-type) which have two membrane-bound cytochrome c molecules in place of subunit II. Quinol oxidases (right) come in several flavors; in the bo3-type, a heme type-o replaces heme a at the binuclear center.
An excellent summary of the liganding of the metal centers in cytochrome oxidase is provided by the PROMISE site cytochrome oxidase pages.
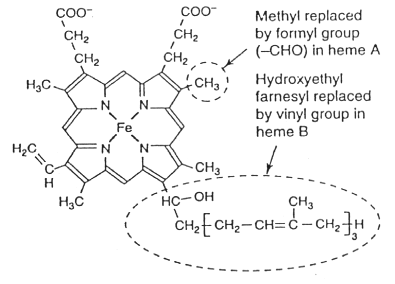
Heme o, showing differences from heme a and heme b.
For cytochrome oxidase, the overall reaction is:
4 ferrocyt c + 4H+N + 4H+N + O2 ==> 4 ferricyt c+ + 2H2O + 4H+P
Since cytochrome c is in the P-phase, 8 charges are transfered from N- to P-phase per oxygen consumed.
The following pictures are from Iwata et al. (1995) Cytochrome Oxidase from Paracoccus denitrificans. Nature 376 660-669; a more complete set can be found on the cytochrome oxidase home page
Click on thumbnails for larger image
Left: Cytochrome oxidase viewed from the side. Subunit I is yellow-green;
Subunit II is purple-red; Subunit III is deep blue. The blue-green subunit
is from the antibody used to aid crystallization of the complex.
Right: Heme a and its protein environment. Note the Fe ligands (His 94
and 413), the ligands to the side-chains of the tetrapyrrole ring, and
the hydrophobic "tail", which anchors the heme group in the protein.
Left: The binuclear center containing heme a3 (heme is pale
blue; Fe is purple-red sphere) and CuB (Cu is blue sphere).
Right: The two Cu atoms of the CuA center in subunit II.
Two views of the redox centers in subunit I.
Left: View from the N-side (periplasm). Heme a is in red, heme a3
is light blue (note European spelling (haem) of heme)
Right: View from the side.
The structure of the beef heart mitochondrial complex has also been solved at a similar resolution, and shows essentially the same structure, but with the many additional small subunits forming membrane helical spans around the periphery of the main catalytic subunits. The cordinates for this structure are now available in the Brookhaven Protein Data Bank (entry 1occ.pdb)
Tsukihara, T., Aoyama, H., Yamashita, E., Tomizaki, T., Yamaguchi, H., Shinzawa-ltoh, K., Nakashima, R., Yaono, R. & Yoshikawa, S. (1996) The Whole Structure of the 13-Subunit Oxidized Cytochrome c Oxidase at 2.5 A. Science 272, 1136-1144.
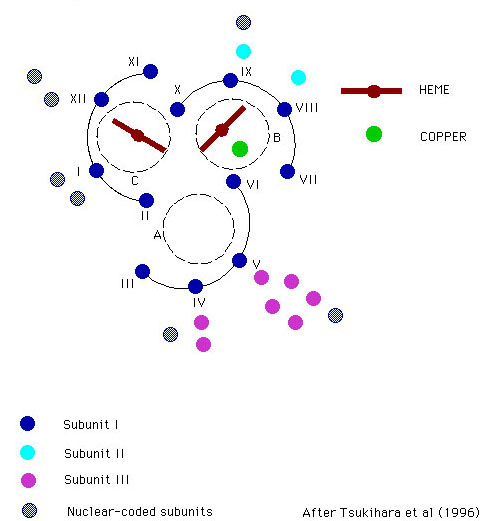
Scheme showing arrangement of helices about the heme centers.
Summary
The crystal structure of bovine heart cytochrome c oxidase at 2.8 A
resolution with an R value of 19.9 percent reveals 13 subunits, each different
from the other, five phosphatidyl ethanolamines, three phosphatidyl glycerols
and two cholates, two hemes A and three copper, one magnesium, and one
zinc. Of 3606 amino acid residues in the dimer, 3560 have been converged
to a reasonable structure by refinement. A hydrogen-bonded system, including
a propionate of a heme A (heme a), part of peptide backbone, and an imidazole
ligand of CuA, could provide an electron transfer pathway between
CuA and heme a. Two possible proton pathways for pumping, each
spanning from the matrix to the cytosolic surfaces, were identified, including
hydrogen bonds, internal cavities likely to contain water molecules, and
structures that could form hydrogen bonds with small possible conformational
change of amino acid side chains. Possible channels for chemical protons
to produce H2O, for removing the produced water, and for O2,
respectively, were identified.
Electron transfer from cytochrome c occurs by electrostatic binding of cyt c to subunit II, followed by electron transfer through CuA center to heme a. Although the docking site and orientation are not known with certainty, the interaction involves a conserved surface of positively charged residue around the heme crevice of cytochrome c, and a complementary surface of acidic residues on subunit II. The electrostatic effect is thought to provided an orienting force sufficient to line up the cytochrome c in the correct orientation during the approach to the docking site. Binding affinity is strongly dependent on ionic stength.
A possible pathway for electron transfer from CuA to heme a has been identified in the structure of the mitochndrial enzyme (see summary above).
Delivery of electrons to the binuclear center is thought to occur to heme a3 via heme a, but the distance from the CuA center to heme a3 is not much greater than the distance from heme a.
The main interest lies in the mechanism of O2-reduction through the binuclear center. The diagrams below shows the dioxygen cycle proposed by Michel (left) or by Wikström (right) for turnover of the binuclear center.
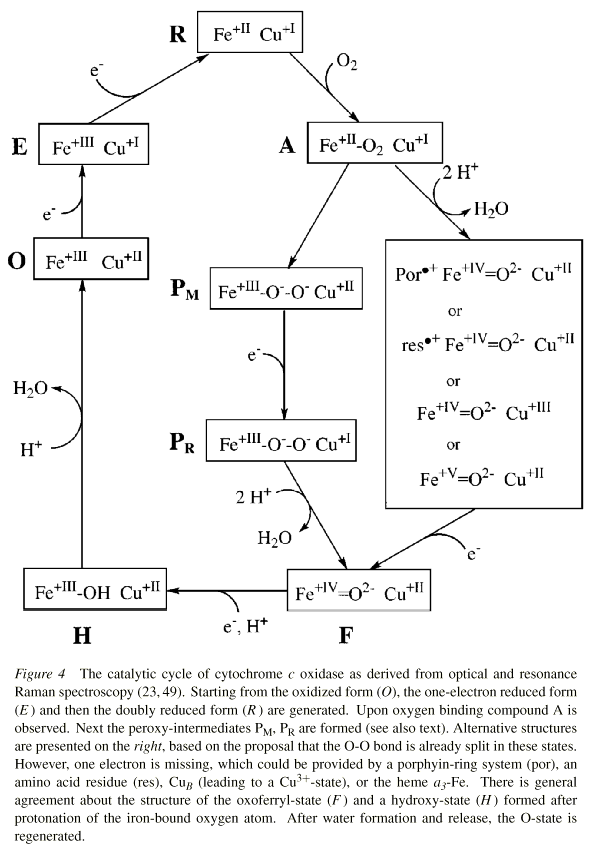
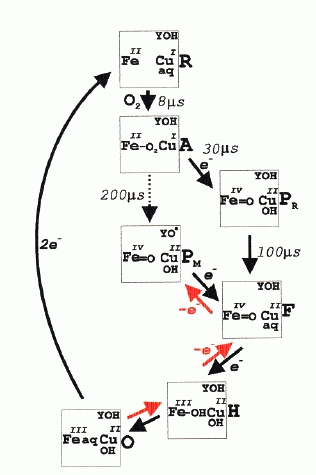
From: (left) H. Michel, J. Behr, A. Harrenga, and A. Kannt (1998) Annu. Rev. Biophys. Biomol. Struct. 27, 329-56; (right) Wikström, M. (2000) Biochemistry, 239, 3515-3519
The binuclear center is represented by its metals, Fe+III/Fe+II for heme a3, Cu+II/Cu+I for CuB.
The scheme is based on a large body of research in which the spectra of the intermediate forms have been characterized, and the kinetics measured using spectrophotometry, and a variety of techniques for rapid mixing and photochemical production of the oxidized form by flash photolysis of CO from the CO-liganded cytochrome oxidase. The scheme shows the 4 scalar protons, but not the protons which are pumped across the membrane; the points of entry of these latter are still controversial. Earlier experiments from Wikstrom had suggested that the work for these extra protons was provided by the reactions in which P is converted to O.
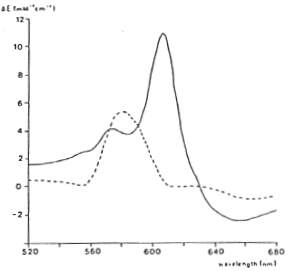
Difference spectra of ferryl (dotted line) and peroxy (solid line) intermediates relative to the oxidized enzyme.
The thermodynamics of the coupling of electron transfer to proton pumping have been determined by experiments from Mårten Wikström's group using intact mitochondria, in which the mitochondrial enzyme was driven in reverse by the proton gradient generated by ATP-hydrolysis.
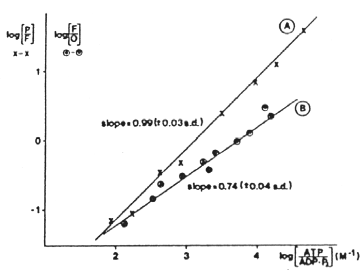
The mitochondria were treated with rotenone, antimycin and myxothiazol to inhibit electron transfer from the respiratory chain, and ferricyanide was added to poise the metal centers in the oxidized (or partly oxidized) state. In A, ferricyanide was present to oxidize all the metal centers. Under these conditions, formation of the P intermediate from O can be driven by the reverse reaction, with the CuA/heme a pair as electron acceptor. In B, a mixture of ferri- and ferrocyanide was present to partly reduce the CuA/heme a pair, and thus limit the availablity of electron acceptors for the reverse reaction. Under these conditions, the reverse reaction forms the F intermediate. By varying the ATP/ADP ratio, the driving force available from ATP-hydrolysis (DG'ATP) could be varied, and the extent of the reaction measured under both conditions. The figure shows how the ratio of reactants for the two redox half-cells varied as a function of the driving force. Since formation of ATP from ADP and phosphate requires 4H+ in intact mitochondria, hydrolysis of ATP pumps 4H+/ATP hydrolysed. In experiment A, the slope of 1 therefore indicates that the transition from P to F involves the transfer of 4H+ across the membrane; the slope of 0.75 in experiment B indicates that the transition from F to O involves the transfer of 3H+ across the membrane. In each case, two of the protons are those involved in reduction of oxygen. Recently, quantitative aspects of this work have been questioned, because the pattern of H+ uptake suggests that some uptake occurs in the reductive phase (see below).
The solution of the structure of cytochrome oxidase has led to much discussion of the mechanism of proton pumping. The figures below are taken from the paper by Iwata et al. (see above), which should be consulted for details. The groups thought to be involved in proton pumping are shown in the figure below. They have been identified by site-directed mutagenesis, mainly in the Gennis lab.
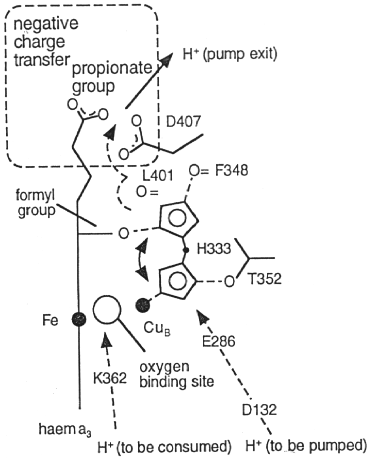
A tentative scheme for proton pumping involving these groups is shown below. Detail aspects of the mechanism are at present controversial; both labs involved in crystallographic studies have demonstrated that the histidine ligand to CuB (H333), which appeared to be mobile in the Iwata structure, is in the liganding position observed in the Yoshikawa structure in both oxidized and reduced enzyme in the absence of azide. It therefore seems unlikely that movement of this group through the sequence of states shown in the scheme below occurs. However, other variants of this scheme with movement linked to other transitions, are not excluded.
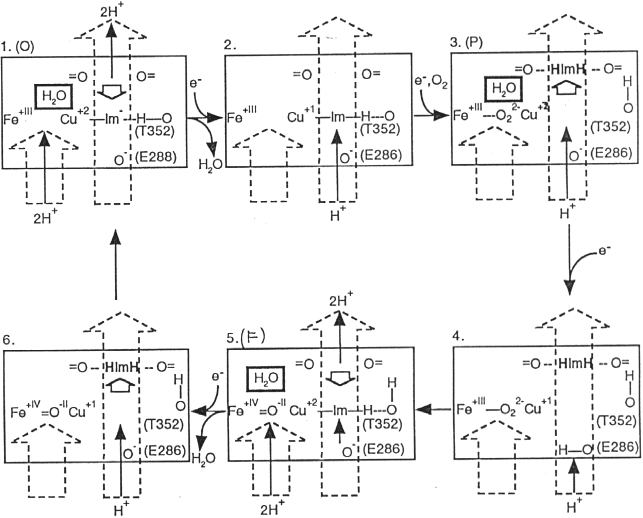
Michel has recently updated these ideas, and challenged Mårten Wikström's earlier conclusions about the points of entry of protons. A somewhat complicated scheme, taken from ref. 2, can be seen below.
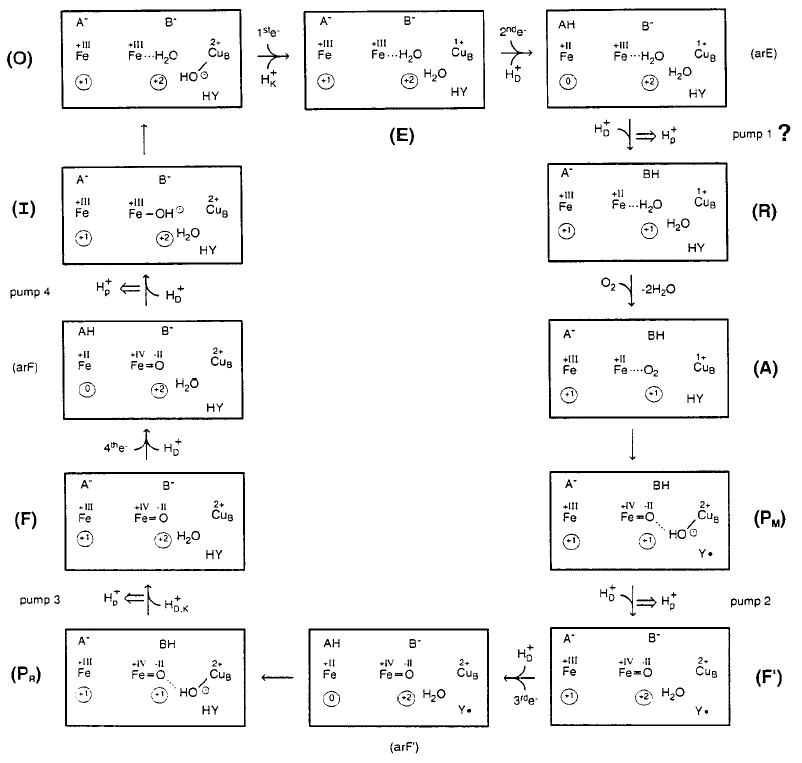
FIGURE 3: Mechanistic model for the catalytic cycle of COX and its coupling to
proton pumping by electrostatic repulsion. The left heme Fe symbol in each
state stands for the heme a Fe atom with +III or +II as its formal
oxidation number. A- stands
for a proton-accepting site at the heme a propionates. The Fe symbol
more to the right stands for the heme a3 iron. A
water molecule (H2O) is bound to it in the O-state. An OH- bound to CuB is
shown. B- stands for a common protonation site near the
heme a3-CuB binuclear
site. For each state the overall net charge at the heme a Fe site
(without A) is indicated within the left-hand circle, and the net charge in the
immediate vicinity of the binuclear site (including HY, without B) is shown
within the right-hand circle. All changes in the boxes are electroneutral. The
reductions of heme a during the E -> R, F' -> PR, and F -> O transitions are charge-compensated by proton uptake via the D pathway; these protons are stored in the
proton-accepting site A. The resulting states are called arE, arF', and arF,
respectively, where ar stands for heme a reduced. The proposal of a
pumping step between arE and R is highly speculative; it might be achieved by
electrostatic repulsion of the proton in the A-site by a proton entering the
B-site via the D-pathway, if proton transfer from the A-site to the B-site is
interrupted. e- stands for electrons
transferred from CuA
to heme a. Protons
taken up via the K-pathway are shown as HK+, those taken up via the D-pathway as HD+and those taken up via the D- or the
K-pathway as HD,K+. Proton pumping is indicated by a double
arrow. Y· stands for the
neutral radical of the H27-H280
cross-link, and Y- for the tyrosinate. For more details see text.
The controversy between Wikstrom and Michel was been played out in recent papers from both groups, which should be consulted for further details:
Reviews: